This article documents content that is no longer in the game
Project Discovery is a "citizen science"—participation in scientific research by the lay community—project within EVE Online. A collaboration between CCP Games, Reyjavik University, the Human Protein Atlas, and Massively Multiplayer Online Science, Project Discovery is used to characterize the structures of cells, using players' identification skills, and will help scientists identify proteins within human cells. Because of this, playing Project Discovery helps scientists understands how human cells are built.
A brief tutorial and introduction to mechanics is present in the game.
In-game, the Sisters of EVE run Project Discovery for capsuleers to assist in identification of Drifter biological samples.
In addition to the above scientific participation and benefits, capsuleers also gain benefits within EVE Online. Each slide submitted results in a small amount of ISK - determined by your accuracy rating, with an accuracy of 50% conferring 50,000 ISK per slide, minus corporation tax - as well as Project Discovery-specific Analysis Kredits with the Sisters of EVE.
The Human Atlas has now ended and the Project Discovery has moved on to exoplanet hunting. To celebrate the success of the first phase, a monument was erected in Lanngisi.
Basic Mechanics
Project Discovery can be accessed using your NeoCom, under the Business tab. Beginning to use it is as simple as pressing this button, then this button and following the tutorial.
Its workflow consists of:
- Completing the Tutorial, if you haven't previously
- Presentation with a new slide
- Clicking on one or more cellular structure buttons, indicating you believe that the fluorescent green is identifying those structures
- Submitting the Image
- Feedback if enough other players have submitted the slide; if not, feedback won't be enabled yet
- - It is at this step that, if consensus of players picks the same options, that your accuracy rating will be adjusted up or down based on which options you pick
- Rewards: ISK, Project Discovery Experience Points, and Sisters of EVE Analysis Kredits (not Loyalty Points)
- - Reward quantities depend upon your accuracy rating
Completing the Tutorial
Before you start playing you will need to complete the tutorial. This teaches you the basic mechanics and what you should be looking for in the slides.
The tutorial will show you:
- How to zoom into the sample image
- How to fix the camera on a certain part of the image
- Where to find the categories
- How to filter the sample image to remove and re-add certain colours
- How to submit your analysis
- Where to find your rank and how to view your progress
- Where to find your accuracy rating
- How to view the tutorial again
You will then be shown ten slides on which a consensus has already been reached. These example slides are of varying difficulty, and you will need to select the category or categories you consider best match the sample slide.
Once you have completed the tutorial, you can start moving on to some other slides. Some of them will already have a consensus and some will not.
The layout of the page
| Image |
Description
|
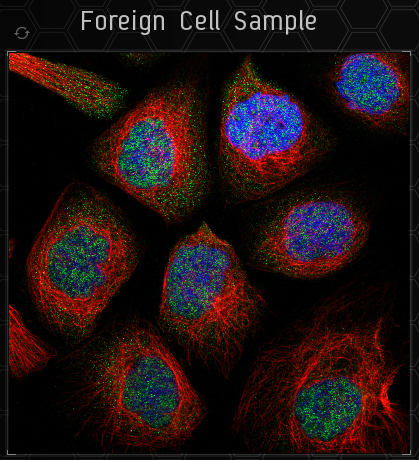 |
This is the sample image. You only need to categorise the green part. To the top left of the sample is a button to load a new sample.
|
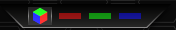 |
Directly below the sample image is the colour filter. You can use this to filter out certain colours to enable you to see the green more easily. You may find it useful to flick back and forth sometimes to see how the colours interact with each other.
|
 |
To the bottom left of the sample image (next to the colour filter) is a checkbox you can use to categorise a sample as 'abnormal'. You should do this when the image doesn't fit within any of the categories, or there is something wrong with the sample image itself.
|
 |
To the right of the sample image are all the possible categories you could select. You are free to select more than one category. If you hove over the categories you will be able to see a description of what each category is, together with several example images showing what this category looks like.
|
 |
At the bottom of the screen is the button to submit your analysis. Only press this when you have selected all the likely categories, as you cannot 'Undo' once it has been submitted.
|
 |
At the top of the window is your current rank. There is also a progress bar underneath which shows you how close you are to obtaining the next rank.
|
 |
Also at the top of the page is your current accuracy. This is based on the samples you have analysed which already have a general consensus. The greater your accuracy, they greater your rewards will be. This is an incentive to be careful when reviewing the samples, because if you get too many wrong your accuracy will start to suffer.
|
 |
Easy to overlook, in the very bottom left corner is the Help button, which enables you to do the tutorial again to re-familiarise yourself with the game.
|
What to Look For
The objective of Project Discovery slides is to identify what cellular structure(s) display a fluorescent green color. These can broadly be classified as within the nucleus, in the cell's cytoplasm, or even outside the cell in the periphery. If none of these are met, consider classifying the slide as not identifiable.
Categories
There are 29 distinct categories. Below is a summary of each and CCP's example image. You can also view three further examples of each by hovering over them in the Project Discovery window.
| Category |
Description |
Image
|
| Nucleus |
The nucleus takes up a big part of the cell volume, and looks like a big, round ball. It overlaps with the blue marker and is easily stained. |
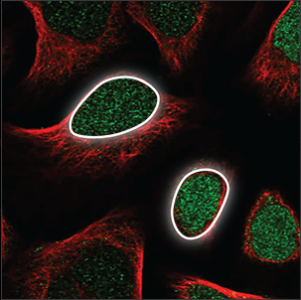
|
| Nucleoplasm |
The nucleoplasm encompasses everything in the nucleus except the nucleoli (a few small, elongated circles) and overlaps with the blue marker. |
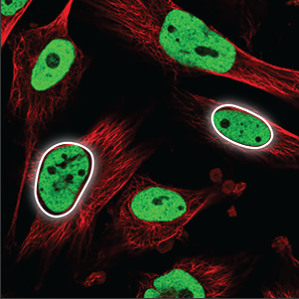
|
| Nuclear bodies (few) |
A few (five or less) distinct spots in the nucleus (blue marker) are stained. |
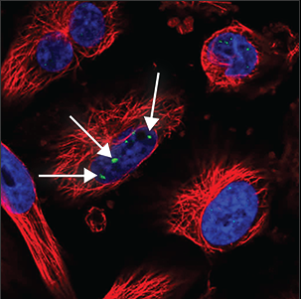
|
| Nuclear bodies (many) |
A high number (more than five) of distinct spots in the nucleus (blue marker) are stained. |
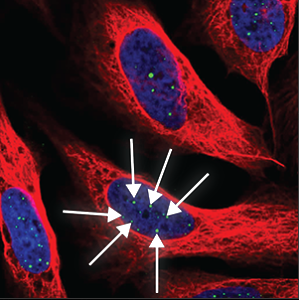
|
| Nuclear speckles |
Distributed throughout the nucleus (blue marker), with their uneven texture they look somewhat like leopard speckles. |
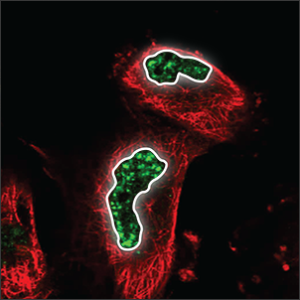
|
| Nucleoli |
A few small, somewhat elongated circles within the nucleus (blue marker), overlapping with holes in the blue marker. |
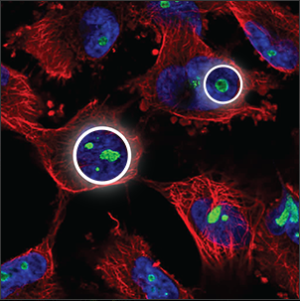
|
| Nucleoli (rim) |
A clear staining of the edge of the nucleoli, which are small, somewhat elongated circles within the nucleus (blue marker), overlapping with holes in the blue marker. |
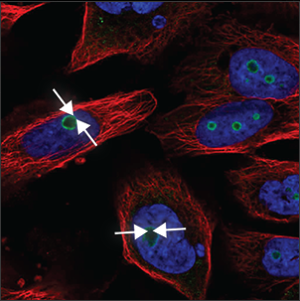
|
| Nucleoli (fibrillar center) |
Clusters of small spots in the nucleoli, which are small, somewhat elongated circles within the nucleus, overlapping with holes in the blue marker. |
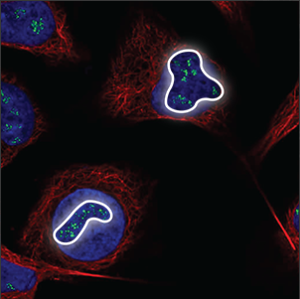
|
| Nuclear membrane |
The nuclear membrane is easy to recognize, the staining forming a thin circle around the nucleus (blue marker). It is also sometimes possible to see the many folds of the nuclear membrane. |
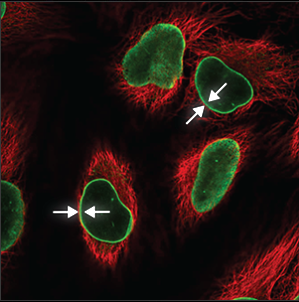
|
| Cytoplasm |
Is seen throughout the whole cell, except in the nucleus (blue marker). The intensity can vary throughout the cell, and is often stronger close to the nucleus. |
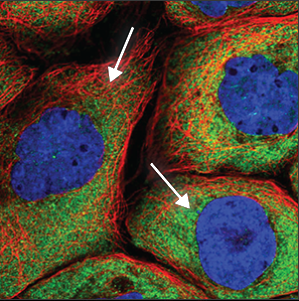
|
| Aggresome |
The aggresome can be seen as a very dense, oval ball next to the nucleus (blue marker). It overlaps with a hole in the microtubules (red marker). |
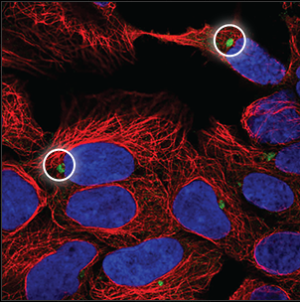
|
| Mitochondria |
A thread like pattern, rather like spaghetti (sometimes long and sometimes chopped up). The mitochondria can be found throughout the cell, starting centrally, close to the nucleus and stretch all the way out to the edges of the cell. |
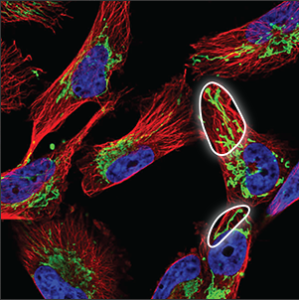
|
| Rods and rings |
Small and distinct rod and ring-like structures, usually only one or a few per cell. |

|
| Cytoskeleton (intermediate filaments) |
Often seen as tangled, rope-like structure throughout the cell. Some are smaller and enclose the nucleus (blue marker), and have a more condensed look. |
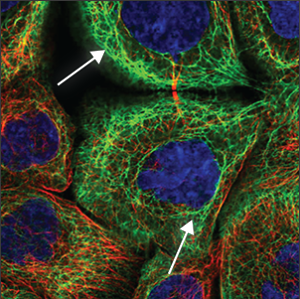
|
| Cytoskeleton (microtubule ends) |
Short strands at the tip or the center of the microtubules (red marker), overlapping perfectly with it. Sometimes only the ends are stained and not the rest of the microtubules. |
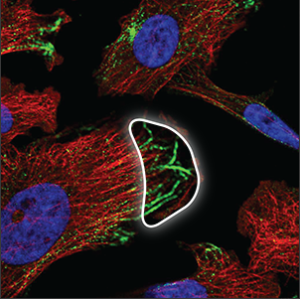
|
| Cytoskeleton (microtubules) |
Thin strands stretching throughout the cell. Overlaps perfectly with the red marker making the colour turn yellow. |
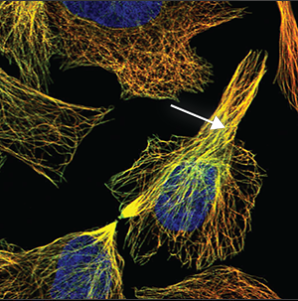
|
| Cytoskeleton (actin filaments) |
Long, very straight and parallel filaments that outline the edges of the cell. |
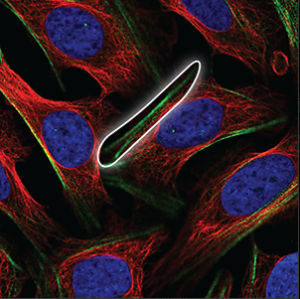
|
| Cytoskeleton (cytokinetic bridge) |
A small structure that can only be seen at the tip between two cells that have almost been separated and moved apart, next to condensed microtubules (red marker). |
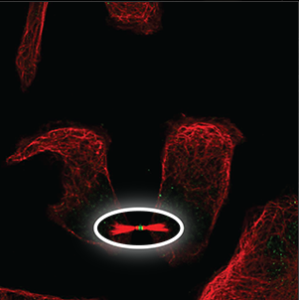
|
| Microtubule organizing center |
A small and somewhat diffuse structure that overlaps with the center of the microtubules (red marker), next to the nucleus (blue marker). |

|
| Centrosome |
Single or double spots which overlaps with the center of the microtubules (red marker), next to the nucleus (blue marker). |
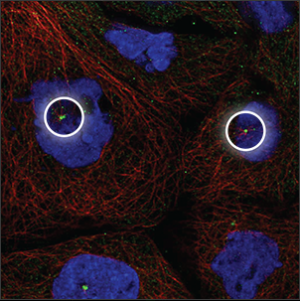
|
| Endoplasmic reticulum |
A symmetrical structure found throughout the cell (excluded from the blue marker). It is a mesh like network that looks somewhat like a spider's web. |
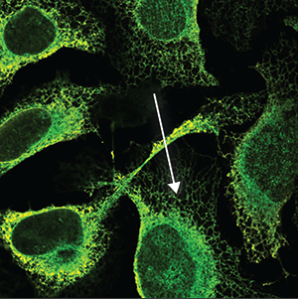
|
| The Golgi apparatus |
The Golgi apparatus is located next to the nucleus, sometimes lying around or above it. It often looks like a collection of small, dense particles that may be donut shaped. |
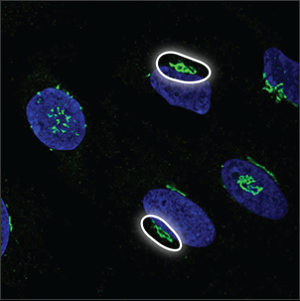
|
| Vesicles |
Small, bright dots that are uniform in size and shape. They are either evenly distributed throughout the cell, or clustering next to the nucleus (blue marker) but never in the nucleus. |
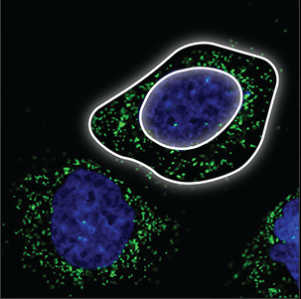
|
| Cell junctions |
Can only be observed when the cells are in contact, at the actual site of cell-cell connections. |
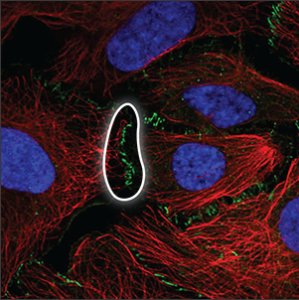
|
| Focal adhesions |
Located under the cell or at the edge of the cell membrane, where the cell is attached to the surface. |
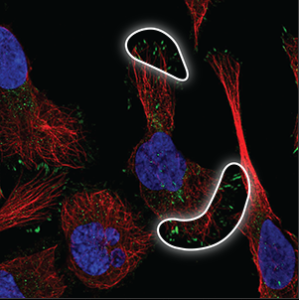
|
| Plasma membrane |
It may appear as a uniform and very flat staining across the entire cell, and can usually be seen clearly outside the red marker. Sometimes only appears as a rim around the cell, and sometimes protrusions are extending from it. |
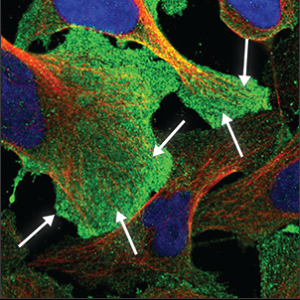
|
| Cell-to-cell variations |
Either varying intensity of the same location in different skills or differences in location between cells. |

|
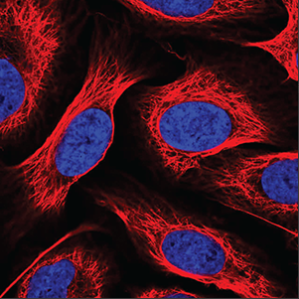
| Negative |
No or extremely weak green staining is seen in the image. |
|
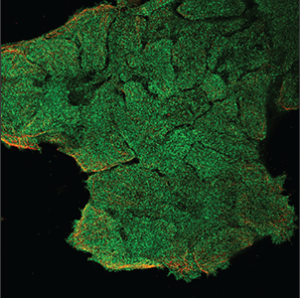
| Unspecific |
No distinguishing patterns can be identified from the staining. All parts of the cell are stained uniformly or staining artifacts are seen outside the cells. |
|
Tips
- Most slides presented to players have 1-2 structures highlighted with fluorescent green, though it's possible that slides may have zero green structures or 3+ green structures.
- Cytoplasm, nucleus, and nucleolus seem to be the most frequent structures identifiable in this batch of images.
- Remember to focus on the fluorescent green color. Consider using the filters at the bottom of the image to hide red and/or blue to make identifying the green region easier.
This image demonstrates a capsuleer selected one element in the Nucleus and one element in the Cytoplasm; a majority of capsuleers agree with the cytoplasm structure choice - hence the green checkmark - but disagree with the nucleus structure choice - hence the red X.

What You Get Out of It
Upon submission of each slide, capsuleers are rewarded with ISK, Project Discovery experience, and Analysis Kredits. The quantity of ISK and Project Discovery experience points are determined by the pilot's accuracy rating: at an accuracy rating of 50%, pilots will accumulate 50 EXP and 50,000 ISK per slide submitted.
Ranks are a method of quantifying the pilot's contribution to Project Discovery. Rank is increased by accumulating experience points.
Analysis Kredits may be spent at Sisters of EVE Loyalty Point stores, but they are not interchangeable with Loyalty Points. Instead, Analysis Kredits are used to purchase new Project Discovery -specific items including boosters and attire.
External Links